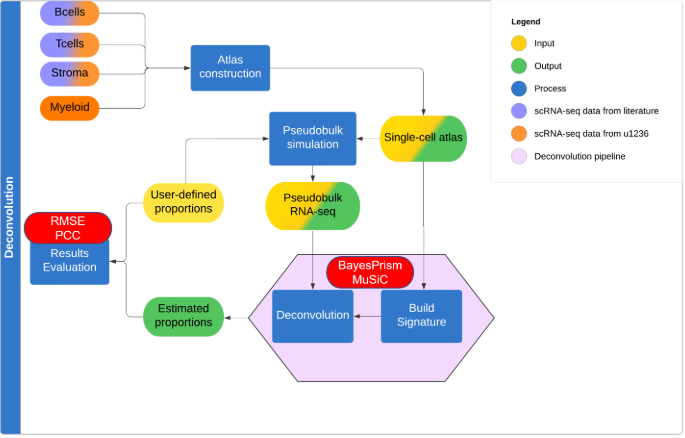
Developed a deconvolution pipeline to estimate cell-type proportions in bulk RNA-seq data using single-cell RNA-seq reference atlases. The project focused on follicular lymphoma and involved conducting a comprehensive literature review, implementing BayesPrism and MuSiC deconvolution methods, and constructing a custom reference atlas. Performance was evaluated using simulations, focusing on cell population granularity, gene signature similarity, and computational efficiency.
Download Report